Atlas Overview: feature space for each development stage
8,432
mRNA Transcripts
3,156
Proteins
283
miRNAs
7 stages
Myeloblast → Neutrophil granulocyte
Comprehensive multi-omics analysis across 7 developmental stages
Hesse, Klein et al., Nature Communications (under revision)
mRNA Transcripts
Proteins
miRNAs
Myeloblast → Neutrophil granulocyte
Explore gene/protein expression patterns across all developmental stages
Launch Interactive ExplorerSearch features • Compare stages • Export results
Interactive versions with underlying data and code
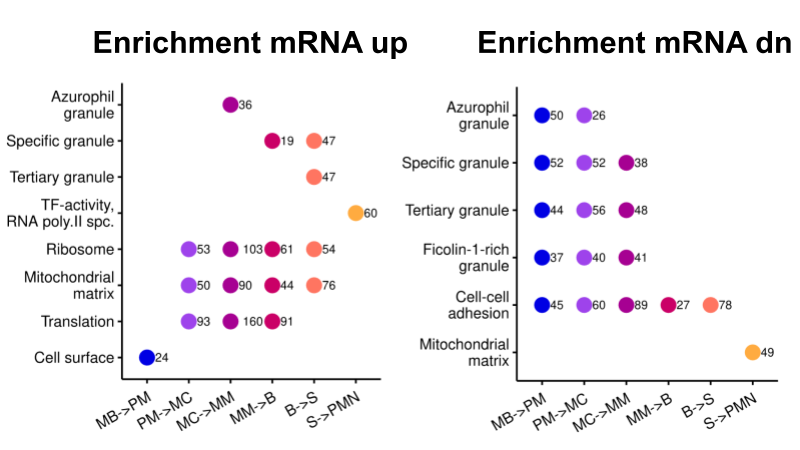
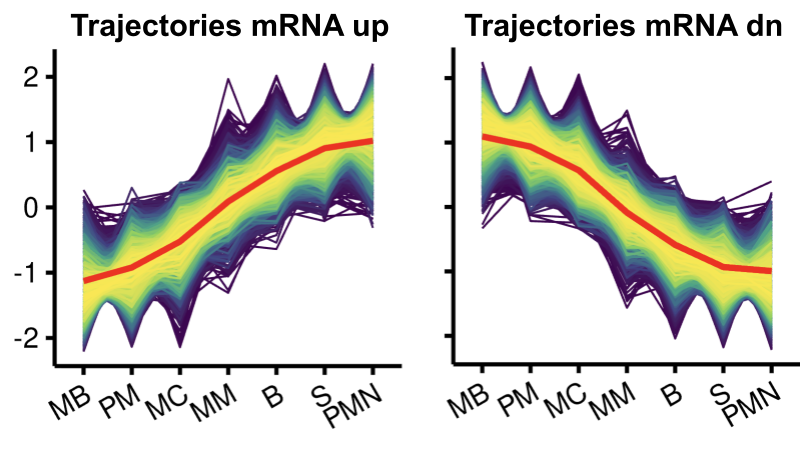
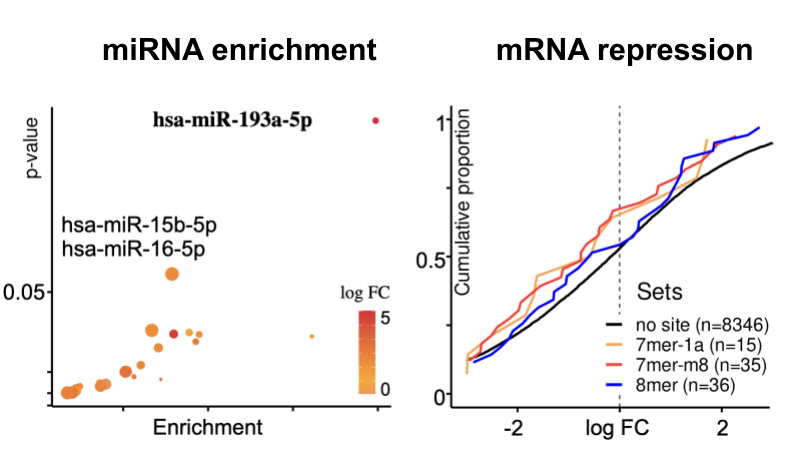
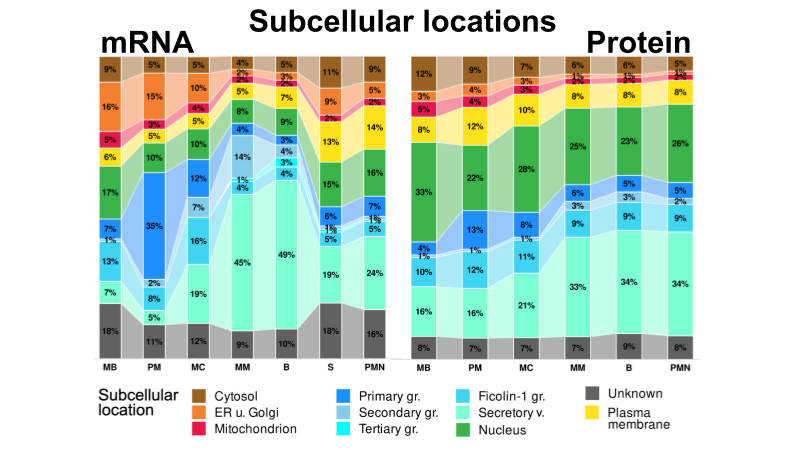
The complete preprint is available on Research Square
View Preprint on Research SquareOpens in a new window
All data formats • VSN, TPM, Log2 normalized • Direct downloads
Complete reproducible analysis pipeline
Complete reproducible analysis pipeline
Nature Communications (under revision)
DOI: Pending publication